Protein Structure Modeling
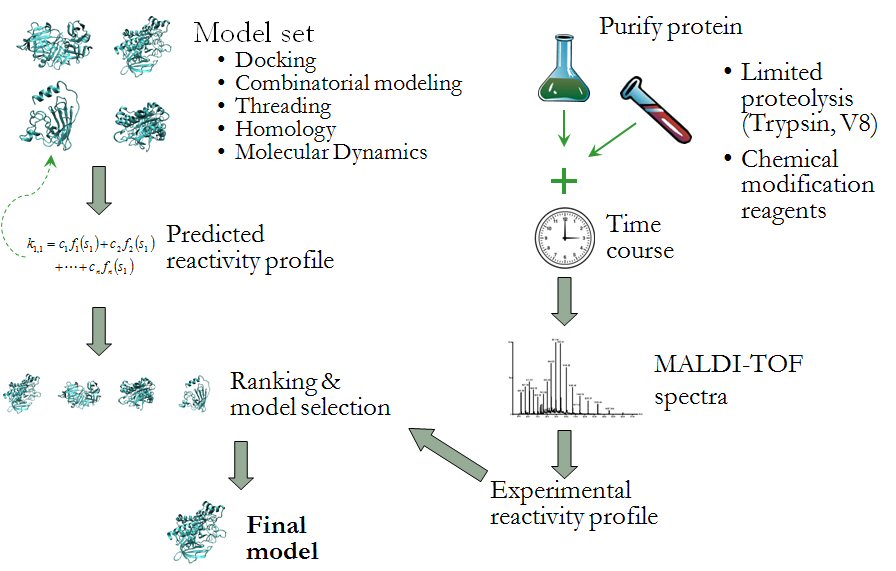
Understanding protein structure and the forces that drive protein folding is one of the most fundamental and challenging problems in biochemistry. We are pursuing a number of projects that explore the determinants of protein structure and improve computational structure prediction methods. Our current areas of investigation include:
- Development of a novel technique for the identification of remote homologs,
- Characterization of secondary structure variability for protein sequences, and
- Hybrid experimental/computational methods for high-confidence prediction of protein tertiary and quaternary structure.
The latter project involves improving the reliability of protein structure prediction algorithms by including experimental information in the model selection process. In collaboration with Dr. Jerry Alter’s lab, (Department of Biochemistry and Molecular Biology, Wright State University) we have developed the computational support for MRAN - Modification Reactivity Analysis (see figure above). Based upon the reaction rate of proteolysis or residue modification reactions, solvent accessibility and other physiochemical properties of specific residues can be estimated. This information can then be used to drive the process of selecting and refining conformational models for further exploration.