Topological Data Analysis of Metabolomic Profiles
A plethora of chemical processes is continuously occurring in our cells at all times, producing chemical products or metabolites. In Metabolomics, these metabolites are measured and characterized with sophisicated chemical detection techniques, such as NMR and LC-MS, giving scientists a snapshot of the chemical ‘state’ of the biological cells, tissues or fluid it was taken from. However, numerous metabolites are often present in a single biological sample, making data analyses particularly challenging. Additionally, substantial variation can be expected within and between individuals, thereby masking the underlying effects driven by disease or treatment.
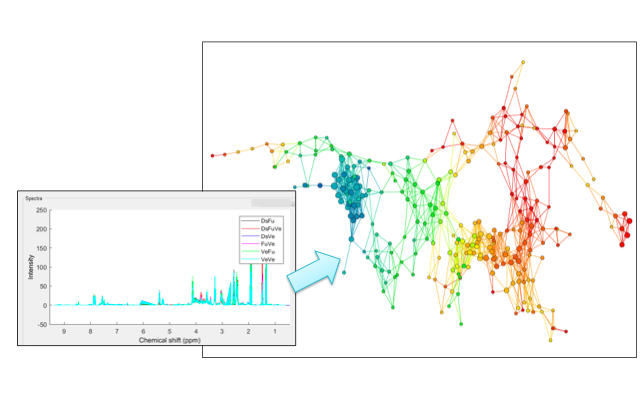
To derive such underlying patterns and trends, we apply Topological Data Analysis (TDA), a powerful data analytic approach that emerged from previous works in Applied Topology and Computational Geometry. In the form of simplicial complices (see right diagram), TDA captures a compressed representation of the topological manifold which describes the trends in the data of interest. Topological patterns, such as flares, loops and voids, may be observed in these simplicial complices. Possible data trends and relationships may be inferred from such patterns, for example, the presence of a unique data cluster, data with cyclical nature, or the boundary conditions for the system from which the data is sampled.
Working with Dr. Nicholas Reo’s Nuclear Magnetic Resonance Lab at Wright State University, we study the utility of TDA in the analysis of NMR spectroscopy of processed biological fluids and tissues (see left diagram).