Understanding Genomic Changes in Cancer Using Machine Learning
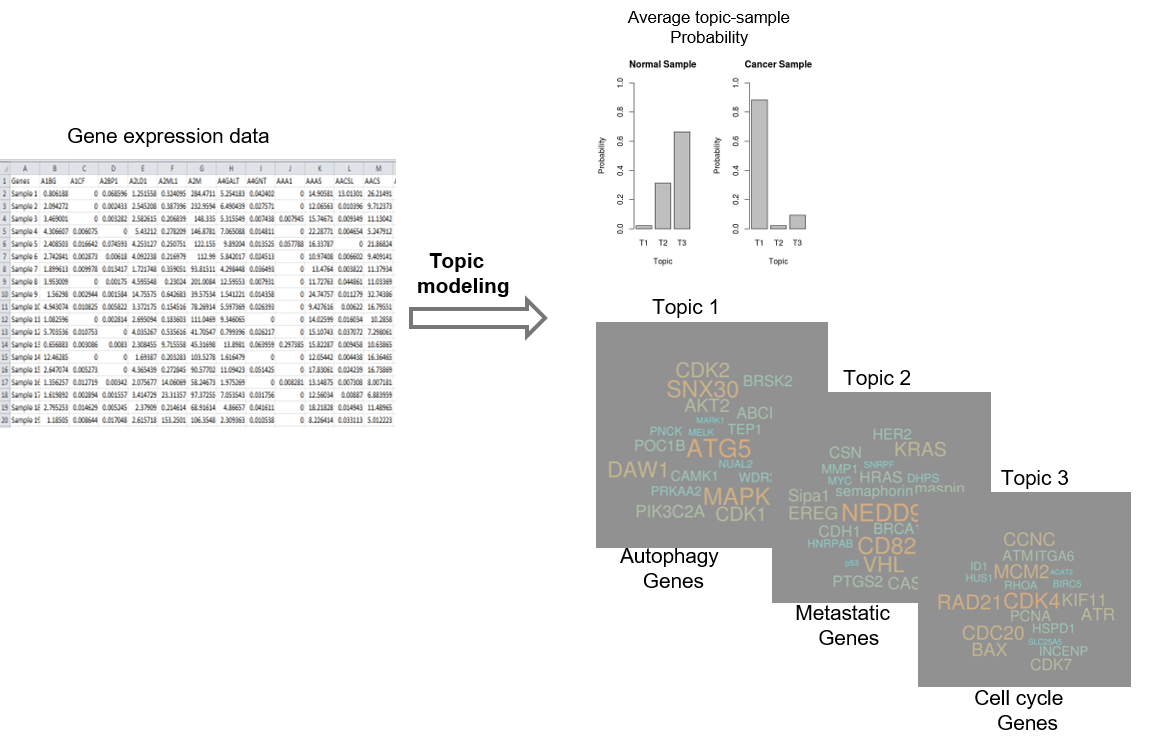
Cancer is a complex disease driven by the combination of various genetic, environmental and lifestyle changes. Understanding the role of differential gene expression in the development of and molecular response to cancer is a complex problem that remains challenging, in part due to the sheer number of genes, gene products, and metabolites involved. In recent years, with the development of high-throughput sequencing, analyzing data is a challenging task particularly because of small sample size and large number of genes (high dimensionality). With such huge dimension space, it appears easy for classic statistical methods to over-fit the data. Further, to select relevant genes involved in different types of cancer is also a challenge. Therefore, methods such as machine learning became popular because of their ability to model non-linear relationships and construct interpretable models. In this project, we investigate the effectiveness of different machine learning approaches on such challenging classification tasks. We have implemented an unsupervised topic model, Latent Dirichlet Allocation (LDA), to mitigate overfitting of high-dimensionality gene expression data and to develop a high-level understanding of the associated genes and pathways involved in breast cancer and lung cancer. Topic modeling is a machine learning approach most commonly employed in text mining to reveal the underlying thematic structure of documents. The basic assumption in topic modeling is that a document can be described as a mixture of latent topics, each with a specific probability distribution over the set of available words. In our application, topics can be synonymous to processes and words are synonymous to genes/transcripts. An important advantage of topic modeling, compared to the existing machine learning algorithms, is its proven ability to handle sparse inputs over extremely large numbers of features in an unsupervised manner.